Week3 : Simple implementation of Parameter estimation using COPASI
Hi all!
Today I will firstly explain what I did this week and discussion of this.
And then I will confirm the task of next week
1. What I did this week
I mainly focus on implementing parameter estimation analysis code using library of COPASI. Completing this task, I can say that fundamental implementation, Simulation, Steady state analysis and parameter estimation is finished and then I can focus on the implementation for users, such as client side UI to change the parameter of analysis.
1.1 Implementation : Server side Java code for analysis of parameter estimation
First of all I implemented the analysis of parameter estimation using COPASI API because I am a little bit familiar with this library. Executing parameter estimation, I supposed that the SBML model and experiment data( normally considered as time-course expression data) are sent from client side. In my implementation the analysis is executed and the result is arranged as JSON format using JSONIC library as I mentioned last week. Then the result is sent to client side by HTTP transfer. In terms of algorithm to execute parameter estimation I selected Leven-Berg Marquadt method in this implementation.
1.2 Implementation : Client side html and JavaScript for analysis of parameter estimation
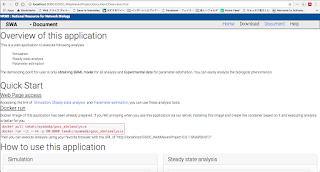
In html file I prepared the form selecting the SBML model and Experiment data. When these data is selected, JavaScript code immediately send these data to Server side and callback function set the result from Server side to form of chart to visualize the result(AJA JSON).
1.3 Demo of this implementation
The SBML model and experiment data exists in my GitHub repository( example/ParameterEstimation). Experiment data is obtained changing original parameter of SBML model using CellDesigner(http://www.celldesigner.org).
1.3.1 Client side screen of parameter estimation
1.3.2 Selecting SBML model
1.3.3 Selecting experiment data
1.3.4 Visualizing the simulation result and experiment data before parameter fitting
1.3.5 Visualizing the simulation result and experiment data after parameter fitting(Scrolling down from Fig.1.3.4 screen)
1.3.6 Confirming the parameter fit is correctly executed
[ Discussion ]
Current implementation is simple but basic approach and visualisation of parameter estimation. In order to let user feel more convenient to analyse, following tasks should be executed.
(1) The colouring of simulation and experiment data of each species.
=> It is so difficult to distinguish each species.
(2) Configuring the time range of simulation
=> You can see the time-range of experiment data is much smaller than simulation result(Fig.1.3.6)
(3) Setting the form to select the parameter which user want to fit.
=> Some parameters can be already identified by other experiment. Current implementation execute parameter estimation for all parameters in model
(4) Setting the form to choose the parameter to execute parameter estimation( ex : the method of parameter estimation , the range of parameter , tolerance etc)
=> Current implementation use only Leven-Berg Marquadt method and the range of parameter is fixed. Initial value of parameter is also fixed using the value of SBML model.
(5) Configuring screen of visualisation
=> the chart of these visualisation result is too big to make user annoying because scroll down is sometimes bothering user to catch overall results.
(6) Adding "Download" form to get the SBML model whose parameter is fitted and the plain text to show the change of parameters( like K : 0.1 -> 0.3)
2 The task of next week following proposal
I will mainly focus on setting the form to select some analysis parameters( ex: Method of parameter estimation etc) for Simulation, Steady state analysis and Parameter estimation and implementing the Server side code to accept these parameters.
Today I will firstly explain what I did this week and discussion of this.
And then I will confirm the task of next week
1. What I did this week
I mainly focus on implementing parameter estimation analysis code using library of COPASI. Completing this task, I can say that fundamental implementation, Simulation, Steady state analysis and parameter estimation is finished and then I can focus on the implementation for users, such as client side UI to change the parameter of analysis.
1.1 Implementation : Server side Java code for analysis of parameter estimation
First of all I implemented the analysis of parameter estimation using COPASI API because I am a little bit familiar with this library. Executing parameter estimation, I supposed that the SBML model and experiment data( normally considered as time-course expression data) are sent from client side. In my implementation the analysis is executed and the result is arranged as JSON format using JSONIC library as I mentioned last week. Then the result is sent to client side by HTTP transfer. In terms of algorithm to execute parameter estimation I selected Leven-Berg Marquadt method in this implementation.
1.2 Implementation : Client side html and JavaScript for analysis of parameter estimation
In html file I prepared the form selecting the SBML model and Experiment data. When these data is selected, JavaScript code immediately send these data to Server side and callback function set the result from Server side to form of chart to visualize the result(AJA JSON).
1.3 Demo of this implementation
The SBML model and experiment data exists in my GitHub repository( example/ParameterEstimation). Experiment data is obtained changing original parameter of SBML model using CellDesigner(http://www.celldesigner.org).
1.3.1 Client side screen of parameter estimation
Fig. 1.3.1 : Client side screen : Firstly user can choose only SBML model
1.3.2 Selecting SBML model
Fig1.3.2 Selecting SBML model
1.3.3 Selecting experiment data
Fig.1.3.3 Selecting experiment data( csv format is considered) : After selecting SBML model , the form to select experiment data open. the format is now considered as csv whose row refers to time and column refers to species and the value is amount or concentration of intended species.
Fig 1.3.4 : Visualization of Simulation result and experiment data before parameter estimation :
The line shows the simulation result and dotted plot is the experiment data. The vertical axis is time and horizontal axis is concentration of species. You can check original parameter can not fit the simulation result to experiment result.
Fig 1.3.5 : Visualization of Simulation result and experiment data after parameter estimation :
The information of figure is totally same as Fig 1.3.4. This figure can be seen scrolling down from Fig1.3.4 screen.
1.3.6 Confirming the parameter fit is correctly executed
Fig.1.3.6 Confirmation of parameter estimation : The simulation result of species A is invisible from Fig1.3.5 and you can confirm the simulation result is actually fit to experiment result.
[ Discussion ]
Current implementation is simple but basic approach and visualisation of parameter estimation. In order to let user feel more convenient to analyse, following tasks should be executed.
(1) The colouring of simulation and experiment data of each species.
=> It is so difficult to distinguish each species.
(2) Configuring the time range of simulation
=> You can see the time-range of experiment data is much smaller than simulation result(Fig.1.3.6)
(3) Setting the form to select the parameter which user want to fit.
=> Some parameters can be already identified by other experiment. Current implementation execute parameter estimation for all parameters in model
(4) Setting the form to choose the parameter to execute parameter estimation( ex : the method of parameter estimation , the range of parameter , tolerance etc)
=> Current implementation use only Leven-Berg Marquadt method and the range of parameter is fixed. Initial value of parameter is also fixed using the value of SBML model.
(5) Configuring screen of visualisation
=> the chart of these visualisation result is too big to make user annoying because scroll down is sometimes bothering user to catch overall results.
(6) Adding "Download" form to get the SBML model whose parameter is fitted and the plain text to show the change of parameters( like K : 0.1 -> 0.3)
2 The task of next week following proposal
I will mainly focus on setting the form to select some analysis parameters( ex: Method of parameter estimation etc) for Simulation, Steady state analysis and Parameter estimation and implementing the Server side code to accept these parameters.








コメント
コメントを投稿